

Scientists at the Hebrew University of Jerusalem and the EPFL Blue Brain Project have developed Neuron_Reduce, a new computational tool that provides the scientific community with a straightforward capability to simplify complex neuron models of any cell type and still faithfully preserve its input-output properties while significantly reducing simulation run-time.
Detailed neuron models consisting of thousands of synapses are key for understanding the computational properties of single neurons and large neuronal networks, and for interpreting experimental results. Simulations of these models are however, computationally expensive (using lots of computing hours), which considerably decreases their utility. For the first time, Scientists at the Hebrew University of Jerusalem and the EPFL Blue Brain Project have formulated a unique analytical approach to the challenge of reducing the complexity of neuron models while retaining their key input/output functions and their computational capabilities.
Reduced models of neurons and networks form a bridge between the highly detailed models capturing minute experimental detail and the simpler models that lend themselves more easily to theoretical interpretation at the cost of missing important details. These bridging models significantly reduce the amount of computation time as well as storage that are needed for detailed neuron models (and the networks they form) and lead to faster simulation time and a larger neuronal circuit that could be simulated.
Neuron_Reduce is a new analytical tool that provides a unique multi-cylindrical reduced model for complex nonlinear neuron models, both in terms of reducing the morphological complexity and reducing computational time. The tool analytically maps the detailed dendritic tree into a reduced multi-cylindrical tree, based on Rall’s cable theory and linear circuit theory. Synapses and ion channels are mapped to the reduced model preserving their transfer impedance to the soma (cell body); synapses with identical transfer impedance are merged into one NEURON process all the while retaining their individual activation times.
“Neuron_Reduce is a significant innovation in analytically modeling dendritic computations,” explains Prof. Idan Segev, David & Inez Myers Professor in Computational Neuroscience and head of the Department of Neurobiology at the Hebrew University of Jerusalem (HUJI). “The analytically reduced model preserves a large set of subthreshold and suprathreshold features of the detailed model, including the identity of individual stem dendrites, their biophysical properties as well as the identity of individual synapses and the variety of excitable ion channels, and it enhances the computational speed of the model by hundreds of times,” Segev says.
A key advantage of the reduction algorithm, also exhibiting its solidity, is that it preserves the magnitude of the transfer impedance from each dendritic location to the soma. Oren Amsalem, Neurobiologist at the HUJI describes why this is so important, “since in linear systems the transfer impedance is reciprocal, Neuron_Reduce also preserves the transfer impedance in the somato-dendritic direction for passive dendritic trees. For example, an injection of current at the soma will result in the same voltage response at the respective dendritic sites in the detailed and reduced models, therefore preserving the bidirectional communication between the soma and the dendrites,” Amsalem says.

An important tool for computational brain science
Another crucial benefit of Neuron_Reduce is that it preserves the identity of individual synapses and their respective dendrites. It also retains specific membrane properties and dendritic nonlinearities, therefore maintaining specific dendritic computations. Furthermore, Neuron_Reduce also conserves the passive cable properties (Rm, Ra, and Cm) of the detailed model, thus preserving synaptic integration and other temporal aspects of the detailed model.
“In the course of applying computational approaches, initially to the mouse brain and ultimately, the human brain, every trick in the box may be needed to make this computationally possible,” says Blue Brain director of computing, Prof. Felix Schürmann. “This includes new generations of computers, innovation in simulation software and more compact modeling formulas such as Neuron_Reduce. Neuron_Reduce can be used not only for more efficient numerical simulations but also for novel neuromorphic hardware adaptations, which today are not able to cope with the cellular complexity present in biophysically detailed tissue models. This reduction method may also allow this gap to be bridged by reducing more detailed models to a representation that is amenable for neuromorphic hardware implementation,” Schürmann says.
“When modeling brain tissue in biophysical detail as we do in the Blue Brain Project, the cost of the simulation in terms of memory requirements or time to solution (due to the number of calculations that need to be performed) matters considerably,” explains Pramod Kumbhar, a High Performance Computing expert at the Blue Brain Project. “Neuron_Reduce is extremely exciting as it opens the path for a novel type of reduced models that crucially maintain important details of the model but possibly run 40-250 times faster. This complements efforts we recently have made on accelerating the simulation technology,” Kumbhar says.
Neuron_Reduce working successfully on a variety of neuron models:
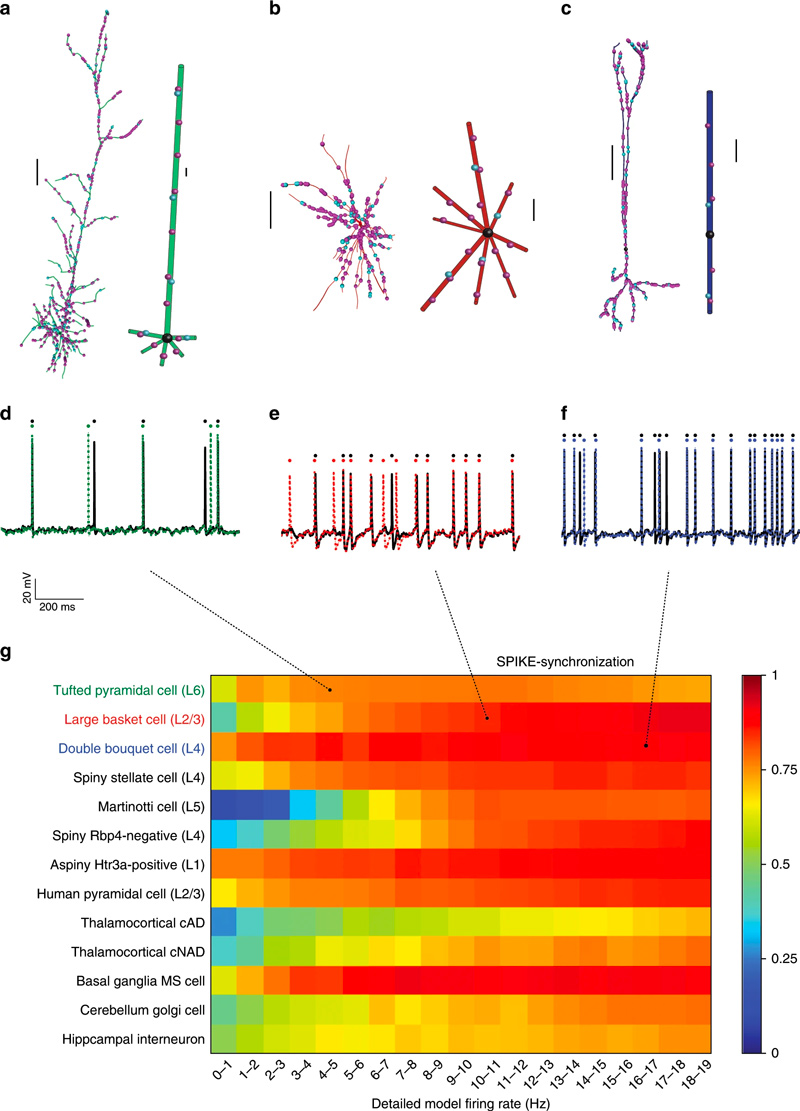
a–c: Detailed models of three somatosensory neurons (left, L6 tufted pyramidal cell in green; middle, L2/3 large basket cell in red; and right, L4 double bouquet cell in blue) and their respective reduced models. Scale bars 100 µm. d–f: Voltage responses to an excitatory synaptic input activated at 1.8, 2.9, and 3.17 Hz, respectively, for both the detailed (black) and the reduced models (corresponding colors). The inhibitory input activation rate was 10 Hz for all models. g: The SPIKE-synchronization index for the 13 detailed versus reduced neuron models. The mean simulation speed-up for the L6 tufted pyramidal cell, L5 Martinotti cell, and L4 spiny stellate cell were 95, 40, and 60, respectively.