

Figuring out what our extinct relatives looked like usually requires fossils – preferably as many as you can get your hands on.
But new research has shown that’s not always the case.
A pioneering study, published in the journal Cell, uses genetic information rather than fossils to predict what our ancient cousins, the Denisovans, might have looked like.
In 2010, scientists sequenced the genome of the previously unknown group of extinct humans from a finger bone found in the Denisova cave in Siberia.
From their genome, we know that Denisovans split off from the Neanderthals around 400,000 years ago, and that at various times they interbred with both the Neanderthals and members of our own species.
But their appearance has remained a mystery.
That’s because aside from the original finger bone, only a handful of teeth and a jawbone have ever been clearly identified as belonging to Denisovans.

To find a work-around, David Gokhman and Liran Carmel at the Hebrew University of Jerusalem in Israel and colleagues turned to the Denisovan genome.
Scientists have long tried to figure out a person’s appearance based on the information contained in their genome. But the sequence of As, Ts, Gs and Cs isn’t enough, says Liran.
“Police would be very happy if they can build an identikit of people based on their DNA sequence, but they simply still cannot do it,” he says. “We took a totally different approach to this.”
Instead of looking at the DNA sequence, Liran and his colleagues searched for patches in the genome that contain tiny chemical blips called methylation. These subtle changes flag gene regulatory regions which dampen a gene’s activity or switch it off entirely.
By comparing DNA methylation patterns in Denisovans to those in modern humans, Neanderthals and chimpanzees, the team could infer which genes are dialled up or down in each group.
“Many anatomical features that change during evolution, the change is attributed to changes in gene regulation, not necessarily changes in gene or DNA sequence,” says Liran.
They then ran these up- or down-regulated genes through a database of human genetic disorders where switching a gene off is known to affect a person’s physical traits.
The team first tested whether their method could accurately predict the physical characteristics of Neanderthals and chimpanzees.
Their method was more than 80% accurate at predicting how a particular trait – such as the thickness of tooth enamel or the width of the face – differs between modern humans and Neanderthals or chimpanzees.
Confident their method was working, they set about predicting what a Denisovan might have looked like.
The team identified 56 skeletal traits where Denisovans differed from modern humans, Neanderthals, or both – 34 of them in the skull.
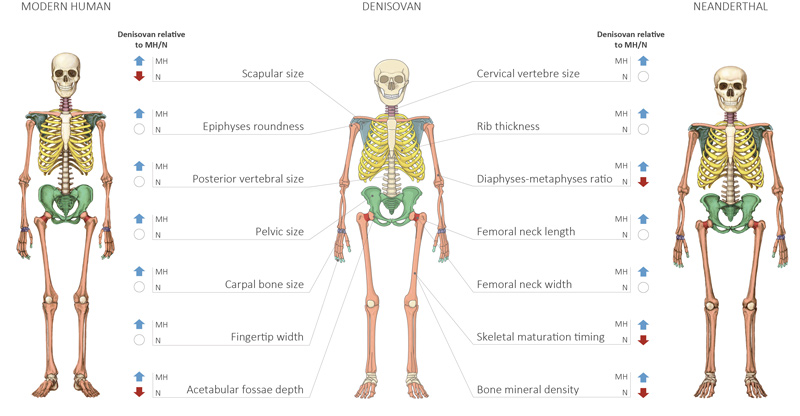
As expected, Denisovans likely looked more similar to Neanderthals than to modern humans, with a large ribcage and pelvis and low forehead, but they probably had a wider skull than both modern humans and Neanderthals.
Their dental arch was also longer, according to the prediction, which fits with the large Denisovan molars that have been found.
While the research was under review, the discovery of a Denisovan mandible was announced. Most features matched those the team predicted.
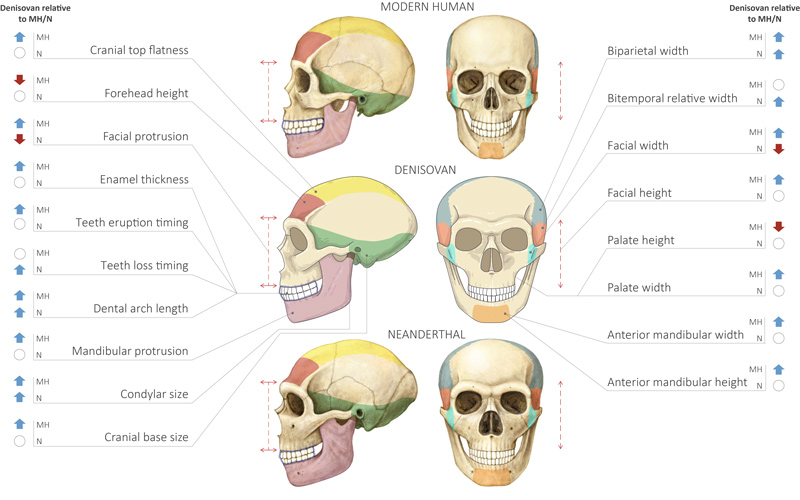
“This was amazing for us, because it was an immediate independent validation of our model,” says Liran.
The findings also lend weight to speculation that two 100,000 year old skulls found in Xuchang in China could belong to Denisovans.
“One of the major properties of these crania is that they were very wide,” says Liran.
Palaeoanthropologist Chris Stringer from the Natural History Museum in London, who was not involved in the study calls it “a pioneering piece of research, which at first glance seems almost like science fiction”.
“This is exciting work, pushing the boundaries of what can be gleaned from ancient genomes,” he says.
The research relies on a complex chain of extrapolations, Stringer notes. Nevertheless, “the results are fully falsifiable by future discoveries”.
